Russell Jones Laboratory
 Cancer and Immunometabolism
Cancer and Immunometabolism
Our mission is to understand what fuels immunity. By investigating the relationship between metabolism, cancer and the immune system, our goal is to translate our findings into the development of novel, innovative approaches for cancer prevention, diagnosis and therapy.
Cancer is essentially a disease in which cells have lost their normal checks on cell proliferation. Cancer cells also have evolved to evade elimination by the immune system, the body’s defense mechanism against foreign invaders. Our research is focused on deciphering the biological perturbations that underlie the development and progression of cancer, with specific interest in studying: 1.) how alterations in cellular metabolism and energy use contribute to tumorigenesis, and 2.) how intrinsic metabolic control shapes immune responses.
Major questions we are investigating include:
- How do cancer cells fuel themselves?
- How do cells adapt to metabolic stress during unchecked growth?
- What are the key metabolic pathways used by T cells to fuel their growth?
- What impact does the tumor microenvironment have on anti-tumor T cell responses?
- Are metabolic pathways good targets for cancer therapy?
Join Our Team
The laboratory of Dr. Russell Jones is searching for postdoctoral fellows and research associates in cancer and immune cell metabolism. Candidates with Ph.D. training in cancer, immunology, mass spectrometry and bioinformatics are encouraged to apply. Visit our career center here.
Recent News & Publications
Learn More
Study reveals how dietary restriction helps fuel cancer-fighting immune cells

Van Andel Institute’s Dr. Russell Jones named to Highly Cited Researchers list

Harnessing the immune system to combat cancer: A Q&A with Postdoctoral Fellow Dr. Michael Dahabieh
Steiner KK … Jones RG, Mogilenko DA, Rathmell JC. 2025. Mitochondrial fatty acid synthesis and MECR regulate CD4+ T cell function and oxidative metabolism. J Immunol 214(5):958–976.
Oswald BM … Jang HJ, Lien EC, Krawczyk CM, Jones RG. 2025. Dietary restriction reprograms CD8+ T cell fate to enhance anti-tumor immunity and immunotherapy responses. Nat Metab.
Jones RG, Simon MC, Kuo CC, Li X, Ho PC. 2025. Conversations at the interface of metabolism and immunity. Mol Cell 85(20):3730-3733.
Longo J, Watson MJ, Williams KS, Sheldon RD, Jones RG. 2025. Nutrient allocation fuels T cell-mediated immunity. Cell Metab.
Kaluba FC, Rogers TJ, Jeong YJ, House RJ, Waldhart A, Sokol KH, Daniels SR, Lee CJ, Longo J, Johnson A, Sartori V, Sheldon RD, Jones RG, Lien EC. 2025. An alternative route for β-hydroxybutyrate metabolism supports cytosolic acetyl-CoA synthesis in cancer cells. Nat Metab.
Our Impact
We’re raising thousands to save millions.
We’re turning hope into action for the millions of people around the world affected by diseases like cancer and Parkinson’s. Find out how you can help us make a difference.
- 141 peer-reviewed papers published in 2025, 74 of which were in high-impact journals
- 15 VAI-SU2C Epigenetics Dream Team clinical trials launched to date
- 10 clinical trials and related projects supported by VAI through the International Linked Clinical Trials Program
Russell Jones, Ph.D.
Chair and Professor, Department of Metabolism and Nutritional Programming
Areas of Expertise
Immunometabolism, cancer metabolism, tumor immunology, metabolomics, diet-genetic interactions in immunology and cancer
Biography
Dr. Russell Jones is a leading expert in the study of cancer metabolism and immunology. As professor and program lead of the Metabolic and Nutritional Programming group at Van Andel Research Institute, his work seeks to uncover how cancer cells fuel themselves through metabolic interactions, with the ultimate goal of developing new cancer therapeutics.
He earned his B.Sc. with honors in Biochemistry and his Ph.D. in Medical Biophysics from the University of Toronto, where he studied in the lab of Dr. Pamela S. Ohashi. After completing a postdoctoral fellowship in the lab of Dr. Craig B. Thompson at University of Pennsylvania in 2008, he accepted a position as an assistant professor in the Department of Physiology and Goodman Cancer Research Centre at McGill University. He was subsequently promoted to associate professor in 2014 and, in 2017, also took on the role of director of the Metabolomics Core Facility at Goodman Cancer Research Centre. He joined Van Andel Research Institute’s Center for Cancer and Cell Biology in 2018 as program lead and a founding member of its Metabolic and Nutritional Programming group. Dr. Jones has earned numerous accolades throughout his career, including a New Investigator Award from the Canadian Institutes of Health Research, the Bernard and Francine Dorval Prize from the Canadian Cancer Society, and several teaching awards at McGill University. He was named a William Dawson Scholar in 2014 and elected to the College of New Scholars, Artists and Scientists of the Royal Society of Canada in 2015. He also serves as a reviewer for a number of journals, including Cell Metabolism, Immunity, Nature, Nature Immunology and Science.
Society memberships
Canadian Society for Immunology
American Association of Immunologists
American Association for Cancer Research
College of New Scholars, Artists and Scientists (Royal Society of Canada)
Cancer and Immunometabolism
Cancer is essentially a disease in which cells have lost their normal checks on cell proliferation. Cancer cells also have evolved to evade elimination by the immune system, the body’s defense mechanism against foreign invaders. Our research is focused on deciphering the biological perturbations that underlie the development and progression of cancer, with specific interest in studying: 1.) how alterations in cellular metabolism and energy use contribute to tumorigenesis, and 2.) how intrinsic metabolic control shapes immune responses.
Major questions we are investigating include:
- How do cancer cells fuel themselves?
- How do cells adapt to metabolic stress during unchecked growth?
- What are the key metabolic pathways used by T cells to fuel their growth?
- What impact does the tumor microenvironment have on anti-tumor T cell responses?
- Are metabolic pathways good targets for cancer therapy?
Immunometabolism
Metabolic control of immune function
One of the fundamental biochemical programs influenced by T cell activation is cellular energy metabolism. Naïve T cells encountering processed antigen and co-stimulatory signals from antigen presenting cells (APCs) switch to a program of anabolic growth and biomass accumulation to promote clonal expansion of antigen-specific T cells. This switch from a quiescent to proliferative state dictates increased demand for ATP and biosynthetic precursors for growth. Activated effector T (Teff) cells reprogram their metabolic activities to meet the increased metabolic demands of cell growth, proliferation and effector function.
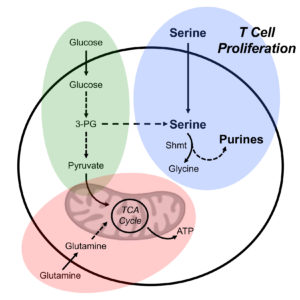
Our research in this area is focused on understanding the key metabolic pathways required by effector T cells for effective immune function. We combine metabolomics with RNA and protein profiling to better understand the essential metabolic nodes that support T cell function. Our current interests are in understanding the impact of serine, glycine and one-carbon (SGOC) metabolism and other branch points from glycolysis and the TCA cycle that impact T cell proliferation and effector function. We also are investigating the impact of nutritional interventions to identify key metabolites that support optimal T cell function in vivo.
Fueling Cancer
Cancer metabolism to drive growth
The ability to engage in increased glycolytic metabolism may be advantageous for growth of cancer cells beyond the ability to rapidly generate energy and suppress apoptosis. This raises a crucial question — what are the metabolic pathways important for growth in proliferating cancer cells?
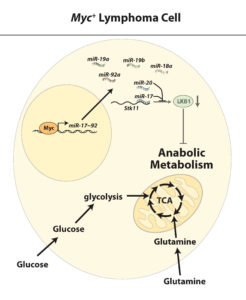
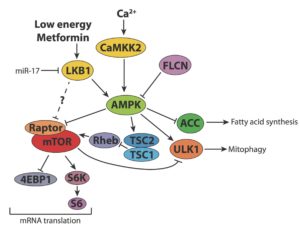
My research group is focused on investigating how signal transduction pathways can redirect metabolic pathways for biosynthesis to enable cell proliferation. Our group focuses on defining the molecular regulators of this process, with particular attention on how the LKB1/AMPK nutrient energy sensing pathway and microRNAs regulate this process.
Surviving lean times
Increased metabolic potential, while supportive of unchecked growth, poses a distinct metabolic challenge for tumor cells. Once a growing tumor outstrips its nutrient supply, it must enact strategies to maintain cellular bioenergetics. Thus, how tumors engage strategies of metabolic adaptation to survive stress may contribute to cancer progression and outcome (i.e., metastasis). Our research in this area has focused on the metabolic regulator AMPK and its role in promoting adaptation to metabolic stress. More recent work has identified pathways that tumor cells use to proliferate when glucose is limiting, which includes co-opting parts of gluconeogenesis to use TCA cycle intermediates for biosynthesis. Our future efforts are focused on identifying novel pathways that mediate cellular adaptation to stress.
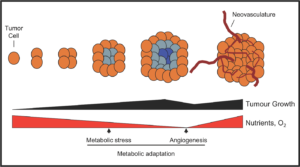
Tumor Immunology
Metabolic competition in the tumor microenvironment
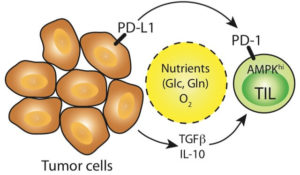
T cells play important roles in cancer immunosurveillance and anti-tumor immunity. Metabolic constraints in the tumor microenvironment can negatively impact anti-tumor T cell responses, in part by competition of tumor infiltrating lymphocytes (TIL) with tumor cells for available nutrients such as glucose. Engagement of receptors such as PD-1 on activated T cells reduces T cell responsiveness and promotes T cell “exhaustion”. Our work focuses on understanding the metabolic constraints that the tumor microenvironment imposes on T cell expansion and function, and whether we can reverse the negative effects of the tumor microenvironment on T cells to enhance anti-tumor immunity.
We also are investigating the impact of metabolic control pathways on the control of inflammation. We have recently found that deregulation of LKB1 in T cells is sufficient to promote gastric tumor formation in mouse models of Peutz Jeghers Syndrome, a cancer predisposition syndrome that arises in patients with LKB1 mutations. This raises the possibility that immune cell-mediated inflammation drives the formation of these cancers. We are actively exploring how LKB1-mediated inflammation control impacts tumor progression.
BIOENERGETICS SPREADSHEET
Download our Bioenergetics Spreadsheet and Instructional PDF to easily convert your seahorse data into bioenergetics figures of merit. This spreadsheet will simplify your analysis and allow you to visually interrogate single or comparative datasets. Our instructional PDF will:
- walk you through entering your data into the spreadsheet
- explain the graphs you will see
- provide instructions for setting up your seahorse experiment to generate data in a way that is easily incorporated into the spreadsheet
The spreadsheet calculations are based off of Mookerjee and Brand’s paper Quantifying Intracellular Rates of Glycolytic and Oxidative ATP Production and Consumption Using Extracellular Flux Measurements and have been updated to reflect his 2019 corrections.
Bioenergetics Spreadsheet
Please fill out the form below to download our Bioenergetics Spreadsheet.
Bioenergetics Spreadsheet
"*" indicates required fields
Selected publications
PubMed | Google Scholar | Mendeley
Steiner KK, Young AC, Patterson AR, Sugiura A, Watson MJ, Preston SEJ, Zhelonkin A, Jennings EQ, Chi C, Heintzman DR, Pahnke AP, Toudji YT, Hatem Z, Madden MZ, Arner EN, Sewell AE, Blount AK, Okparaugo R, Fallman E, Krystofiak ES, Sheldon RD, Gibson-Corley KN, Voss K, Nowinski SM, Jones RG, Mogilenko DA, Rathmell JC. 2025. Mitochondrial fatty acid synthesis and MECR regulate CD4+ T cell function and oxidative metabolism. J Immunol 214(5):958–976.
Oswald BM, DeCamp LM, Longo J, Dahabieh MS, Bunda N, Johnson BK, Watson MJ, Ma S, Preston SEJ, Sheldon R, Vincent MP, Ellis AE, Sopper-Hopper MT, Isaguirre C, Kamarudin D, Shen H, Williams KS, Crawford PA, Kaech S, Jang HJ, Lien EC, Krawczyk CM, Jones RG. 2025. Dietary restriction reprograms CD8+ T cell fate to enhance anti-tumor immunity and immunotherapy responses. Nat Metab.
Jones RG, Simon MC, Kuo CC, Li X, Ho PC. 2025. Conversations at the interface of metabolism and immunity. Mol Cell 85(20):3730-3733.
Longo J, Watson MJ, Williams KS, Sheldon RD, Jones RG. 2025. Nutrient allocation fuels T cell-mediated immunity. Cell Metab.
Kaluba FC, Rogers TJ, Jeong YJ, House RJ, Waldhart A, Sokol KH, Daniels SR, Lee CJ, Longo J, Johnson A, Sartori V, Sheldon RD, Jones RG, Lien EC. 2025. An alternative route for β-hydroxybutyrate metabolism supports cytosolic acetyl-CoA synthesis in cancer cells. Nat Metab.
Longo J, DeCamp LM, Oswald BM, Teis R, Reyes-Oliveras A, Dahabieh MS, Ellis AE, Vincent MP, Damico H, Gallik KL, Foy NM, Compton SE, Capan CD, Williams KS, Esquibel CR, Madaj ZB, Lee H, Roy DG, Krawczyk CM, Haab BB, Sheldon RD, Jones RG. 2025. Glucose-dependent glycosphingolipid biosynthesis fuels CD8+ T cell function and tumor control. Cell Metab.
Dahabieh MS, DeCamp LM, Oswald BM, Kitchen-Goosen SM, Fu Z, Vos M, Compton SE, Longo J, Foy NM, Williams KS, Ellis AE, Johnson A, Sodiya I, Vincent M, Lee H, Yao C, Wu T, Sheldon RD, Krawczyk CM, Jones RG. 2025. The prostacyclin receptor PTGIR is a NRF2-dependent regulator of CD8+ T cell exhaustion. Nat Immunol.
*Featured in a News & Views article
Compton SE, DeCamp LM, Oswald BM, Kitchen-Goosen SM, Lau KH, Fillinger R, Dahabieh MS, Vander Ark A, Krawczyk CM, Jones RG. 2025. IL-17 links the tumor suppressor LKB1 to gastrointestinal inflammation and polyposis. Sci Adv 11(25).
Ma S, Dahabieh MS, Mann TH, Zhao S, McDonald B, Song W, Chung K, Farsakoglu Y, Garcia-Rivera L, Hoffmann FA, Xu S, Du VY, Chen D, Furgiuele J, LaPorta M, Jacobs E, DeCamp LM,Oswald BM, Sheldon RD, Ellis AE, Jang C, Jones R, Kaech SM. 2024. Nutrient-driven histone code determines exhausted CD8+ T cell fates. Science.
Kaymak I, Watson MJ, Oswald BM, Ma S, Johnson BK, DeCamp LM, Mabvakure BM, Luda KM, Ma EH, Lau KH, Fu Z, Muhire B, Kitchen-Goosen SM, VanderArk A, Dahabieh MS, Samborska B, Vos M, Shen H, Fan ZP, Roddy TP, Kingsbury GA, Sousa CM, Krawczyk CM, Williams KS, Sheldon RD, Kaech SM, Roy DG, Jones R. 2024. ACLY and ACSS2 link nutrient-dependent chromatin accessibility to regulation of CD8 T cell effector responses. J Exp Med 221(9).
Drougard A, Ma EH, Wegert V, Sheldon R, Panzeri I, Vatsa N, Apostle S, Facnocchi L, Schaf J, Gossens K, Völker J, Pang S, Bremser A, Dror E, Giacona F, Sagar, Henderson MX, Prinz M, Jones RG, Pospisilik JA. 2024. An acute microglial metabolic response controls metabolism and improves memory. eLife.
Guak H, Weiland M, Vander Ark A, Zhai L, Lau K, Corrado M, Davidson P, Asiedu E, Mabvakure B, Compton S, DeCamp L, Scullion CA, Jones RG, Nowinski S, Krawczyk C. 2024. Transcriptional programming mediated by the histone demethylase KDM5C regulate dendritic cell population heterogeneity and function. Cell Rep 43(8):114506.
Ma EH, Dahabieh MS, DeCamp LM, Kaymak I, Kitchen-Goosen SM, Oswald BM, Longo J, Roy DG, Verway MJ, Johnson RM, Samborska B, Duimstra LR, Scullion LR, Steadman M, Vos M, Roddy TP, Krawczyk CM, Williams KS, Sheldon RD, Jones RG. 2024. 13C metabolite tracing reveals glutamine and acetate as critical in vivo fuels for CD8 T. Sci Adv 10(22).
Luda KM, Longo J, Kitchen-Goosen SM, Duimstra LR, Ma EH, Watson MJ, Oswald BM, Fu Z, Madaj Z, Kupai A, Dickson BM, DeCamp LM, Dahabieh MS, Compton SE, Teis R, Kaymak I, Lau KH, Kelly DP, Puchalska P, Williams KS, Krawczyk CM, Lévesque D, Boisvert FM, Sheldon RD, Rothbart SB, Crawford PA, Jones RG. 2023. Ketolysis drives CD8+ T cell effector function through effects on histone acetylation. Immunity.
Compton SE, Kitchen-Goosen SM, DeCamp LM, Lau KH, Mabvakure, Vos M, Williams KS, Wong KK, Shi X, Rothbart SB, Krawczyk CK, Jones RG. 2023. LKB1 controls inflammatory potential through CRTC2-dependent histone acetylation. Mol Cell.
Madaj ZB, Dahabieh MS, Kamalumpundi V, Muhire B, Pettinga J, Siwicki RA, Ellis AE, Isaguirre C, Escobar Galvis ML, DeCamp L, Jones RG, Givan SA, Adams M, Sheldon RD. 2023. Prior metabolite extraction fully preserves RNAseq quality and enables integrative multi-omics analysis of liver metabolic response to viral infection. RNA Biol 20(1).
Zhu B, Wei X, Narasimhan H, Qian W, Zhang R, Cheon IS, Wu Y, Li C, Jones RG, Kaplan MH, Vassallo RA, Braciale TJ, Somerville L, Colca JR, Pandey A, Jackson PEH, Mann BJ, Krawczyk CM, Sturek JM, Sun J. 2023. Inhibition of the mitochondrial pyruvate carrier simultaneously mitigates hyperinflammation and hyperglycemia in COVID-19. Sci Immunol 8(82).
Kilgour MK, MacPherson S, Zacharias LG, LeBlanc J, Babinszky S, Kowalchuk G, Parks S, Sheldon RD, Jones RG, DeBarardinis RH, Hamilton PT, Watson PH, Lum JJ. 2022. Principles of reproducible metabolite profiling of enriched lymphocytes in tumors and ascites from human ovarian cancer. Nat Protoc 17(11):2668–2698.
Longo J, Watson MJ, Vos MJ, Williams KS, Jones RG. 2022. PYGBacking on glycogen metabolism to fuel early memory t cell recall responses. Mol Cell 82(16):2918–2921.
Samborska B, Roy DG, Rahbani JF, Hussain MF, Ma EH, Jones RG, Kazak L. 2022. Creatine transport and creatine kinase activity is required for CD8 + T cell immunity. Cell Rep 38(9):110446.
Stetzik L, Mercado G, Smith L, George S, Quansah E, Luda K, Schulz E, Meyerdirk L, Lindquist A, Bergsma A, Jones RG, Brundin L, Henderson MX, Pospisilik JA, Brundin P. 2022. A novel automated morphological analysis of IBA1+ microglia using a deep learning assisted model. Front Cell Neurosci 16:944875.
Guak H, Sheldon RD, Beddows I, Vander Ark A, Weiland MJ, Shen H, Jones RG, St-Pierre J, Ma EH, Krawczyk CM. 2022. PGC-1B maintains mitochondrial metabolism and restrains inflammatory gene expression. Sci Rep12:16028.
Kaymak I, Luda KM, Duimstra LR, Ma EH, Longo J, Dahabieh MS, Faubert B, Oswald BM, Watson MJ, Kitchen-Goosen SM, DeCamp LM, Compton SE, Fu Z, DeBerardinis RJ, Williams KS, Sheldon RD, Jones RG. 2022. Carbon source availability drives nutrient utilization in CD8+ T cells. Cell Metab.
Sheldon RD, Ma EH, DeCamp LM, Williams KS, Jones RG. 2021. Interrogating in vivo T-cell metabolism in mice using stable isotope labeling metabolomic and rapid cell sorting. Nat Proto.
Izreig S, Gariepy A, Kaymak I, Bridges HR, Donayo AO, Bridon G, DeCamp LM, Kitchen-Goosen SM, Avizonis D, Sheldon RD, Laister R, Minden MD, Johnson NA, Duchaine TF, Rudoltz MS, Yoo S, Pollak MN, Williams KS, Jones RG. 2020. Repression of LKB1 by miR-17~92 sensitizes MYC-dependent lymphoma to biguanide treatment. Cell Rep Med.
Cordeiro B, Jeon P, Boukhaled GM, Carrado M, Lapohos O, Roy DG, Williams KS, Jones RG, Gruenheid S, Sagan SM, Krawczyk CM. 2020. MicroRNA-9 fine-tunes dendritic cell function by suppressing negative regulators in a cell-type-specific manner. Cell Rep 31(4).
Roy DG*, Chen J*, Mamane V, Ma EH, Muhire BM, Sheldon RD, Shorstova T, Koning R, Johnson RM, Esaulova E, Williams KS, Hayes S, Steadman M, Samborska B, Swain A, Daigneault A, Chubukov V, Roddy TP, Foulkes W, Pospisilik JA, Dourgeois-Daigneualt, Artyomov MN, Witcher M, Krawczyk CM, Larochelle C, Jones RG. 2020. Methionine metabolism shapes helper T helper cell responses through regulation of epigenetic reprogramming. Cell Metab 31(2):250–266.
*Co-first authors
Highlighted in a preview in Cell Metabolism
EH Ma, Verway MJ, Johnson RM, Roy DG, Steadman M, Hayes S, Williams KS, Sheldon RD, Samborska B, Kosinski PA, Kim H, Griss T, Faubert B, Condotta SA, Krawczyk CM, DeBerardinis RJ, Marsh K, Richer MJ, Chubukov V, Roddy T, Jones RG. 2019. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity.
Saibil SD*, St. Paul *, Laister RC, Garcia-Batres CR, Israni-Winger K, Elford AR, Grimshaw N, Robert-Tissot C, Roy DG, Jones RG, Nguyen LT, Ohashi PS. 2019. Activation of peroxisome proliferator activated receptors α and δ synergizes with inflammatory signals to enhance adoptive cell therapy. Cancer Res 79(3):445–451.
Poffenberger MC, Metcalfe-Roach A, Aguilar E, Chen J, Hsu BE, Wong AH, Johnson RM, Flynn B, Samborska B, Ma EH, Gravel SP, Tonelli L, Devorkin L, Kim P, Hall A, Izreig S, Loginicheva E, Beauchemin N, Siegel PM, Artyomov MN, Lum JJ, Zogopoulos G, Blagih J, Jones RG. 2018. LKB1 deficiency in T cells promotes the development of gastrointestinal polyposis. Science 361(6400):406–411.
Raud B, Roy DG, Divakaruni AS, Tarasenko TN, Franke R, Ma EH, Samborska B, Hsieh WY, Wong AH, Stüve P, Arnold-Schrauf C, Guderian M, Lochner M, Rampertaap S, Romito K, Monsale J, Brönstrup M, Bensinger SJ, Murphy AN, McGuire PJ, Jones RG, Sparwasser T, Berod L. 2018. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab.
Guak H, Al Habyan S, Ma EH, Aldossary H, Al-Masri M, Won SY, Ying T, Fixman ED, Jones RG, McCaffrey LM, Krawczyk CM. 2018. Glycolytic metabolism is essential for CCR7 oligomerization and dendritic cell migration. Nat Commun 9(1):2463.
Hall DT, Griss T, Ma JF, Sanchez BJ, Sadek J, Tremblay AMK, Mubaid S, Omer A, Ford RJ, Bedard N, Pause A, Wing SS, Di Marco S, Steinberg GR, Jones RG, Gallouzi IE. 2018. The AMPK agonist 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), but not metformin, prevents inflammation-associated cachectic muscle wasting. EMBO Mol Med 10(7).
Tzelepis F, Blagih J, Khan N, Gillard J, Mendonca L, Roy DG, Ma EH, Joubert P, Jones RG, Divangahi M. 2018. Mitochondrial cyclophilin D regulates T cell metabolic responses and disease tolerance to tuberculosis. Sci Immunol 3(23).
Raud B, McGuire PJ, Jones RG, Sparwasser T, Berod L. 2018. Fatty acid metabolism in CD8+ T cell memory: Challenging current concepts. Immunol Rev 283(1):213-231.
Robichaud N, Hsu BE, Istomine R, Alvarez F, Blagih J, Ma EH, Morales SV, Dai DL, Li G, Souleimanova M, Guo Q, Del Rincon SV, Miller WH Jr, Ramón Y Cajal S, Park M, Jones RG, Piccirillo CA, Siegel PM, Sonenberg N. 2018. Translational control in the tumor microenvironment promotes lung metastasis: Phosphorylation of eIF4E in neutrophils. Proc Natl Acad Sci U S A 115(10):E2202-E2209.
Rabinovitch R, Samborska B, Faubert B, Gravel S-P, Andrzejewski S, Ma EH, Raissi TC, Pause A, St.-Pierre J, Jones RG. 2017. AMPK maintains cellular metabolic homeostasis through regulation of mitochondrial reactive oxygen species. Cell Rep 21(1):1–9.
Jones RG, Pearce EJ. 2017. MenTORing immunity: mTOR signaling in the development and function of tissue resident immune cells. Immunity 46(5):730–742.
Ma EH, Poffenberger MC, Wong AH-T, Jones RG. 2017. The role of AMPK in T cell metabolism and function. Curr Opin Immunol 46:45–52.
Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer M, Verway MJ, Raissi TC, Tsui H, Boukhaled G, da Costa SH, Frezza C, Krawczyk CM, Richer MJ, Hess C, Jones RG. 2017. Serine is an essential metabolite for effector T cell expansion. Cell Metab 25(2):345–357.
Carbonneau MM, Gagné L, Lalonde ME, Germain MA, Motorina A, Guiot MC, Secco B, Vincent EE, Tumber A, Hulea L, Bergeman J, Oppermann U, Jones RG, Laplante M, Topisirovic I, Petrecca K, Huot MÉ, Mallette FA. 2016. The oncometabolite 2-hydroxyglutarate activates the mTOR signaling pathway. Nat Commun 7:12700.
Lehuédé C, Dupuy F, Rabinovitch R, Jones RG, Siegel PM. 2016. Metabolic plasticity as a determinant of tumour growth and metastasis. Cancer Res 76(18):5201–5208.
Izreig S, Samborska B, Johnson RM, Sergushichev A, Ma EH, Lussier C, Donayo AO, Sagan SM, Vincent EE, Artyomov MN, Duchaine TF, Jones RG. 2016. The miR-17~92 microRNA cluster is a global regulator of tumor metabolism. Cell Rep 16(7):1915–1928.
Rao VT, Khan D, Jones RG, Nakamura DS, Kennedy TE, Cui QL, Rone MB, Healy LM, Watson R, Ghosh S, Antel JP. 2016. Potential benefit of the charge-stabilized nanostructure saline RNS60 for myelin maintenance and repair. Sci Rep 6:30020.
Lampropoulou V, Sergushichev AA, Bambouskova M, Nair S, Vincent EE, Loginicheva E, Cervantes-Barragan L, Ma X, Huang SC, Griss T, Weinheimer CJ, Khader S, Randolph GJ, Pearce EJ, Jones RG, Diwan A, Diamond MS, Artyomov MN. 2016. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab 24(1):158–166.
Sergushichev AA, Loboda AA, Jha AK, Vincent EE, Driggers EM, Jones RG, Pearce EJ, Artyomov MN. 2016. GAM: a web-service for integrated transcriptional and metabolic network analysis. Nucleic Acids Res 44(W1):W194–200.
Rone MB, Cui QL, Fang J, Wang LC, Zhang J, Khan D, Bedard M, Almazan G, Ludwin SK, Jones RG, Kennedy TE, Antel JP. 2016. Oligodendrogliopathy in multiple sclerosis: Low glycolytic metabolic rate promotes oligodendrocyte survival. J. Neurosci 36(17):4698–4707.
Balmer ML, Ma EH, Bantug GR, Grählert J, Pfister S, Glatter T, Jauch A, Dimeloe S, Slack E, Dehio PG, Krzyzaniak MA, King CG, Burgener A-V, Fischer M, Develioglu L, Belle R, Recher M, Bonilla WV, Macpherson AJ, Hapfelmeier S, Jones RG, Hess C. 2016. Memory CD8+ T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44(6):1312–1324.
Ma EH, Jones RG. 2016. (TORC)ing up purine biosynthesis. Science 351(6274):670–671.
Griss T, Vincent EE, Egnatchik R, Chen J, Ma EH, Faubert B, Viollet B, DeBerardinis RJ, Jones RG. 2015. Metformin suppresses cancer cell proliferation by suppressing mitochondrial-dependent biosynthesis. PLoS Biol 13(12):e1002309.
Ramiscal RR, Parish IA, Lee-Young RS, Babon JJ, Blagih J, Pratama A, Martin J, Hawley N, Cappello JY, Nieto PF, Ellyard JI, Kershaw NJ, Sweet RA, Goodnow CC, Jones RG, Febbraio MA, Vinuesa CG, Athanasopoulos V. 2015. Attenuation of AMPK signaling by ROQUIN promotes T follicular helper cell formation. eLife e08698.
Bott AJ, Peng I-C, Fan Y, Faubert B, Zhao L, Li J, Neidler S, Sun Y, Jaber N, Krokowski D, Lu W, Pan JA, Powers S, Rabinowitz J, Hatzoglou M, Murphy DJ, Jones RG, Wu S, Girnun G, Zong W-X. 2015. Oncogenic Myc induces expression of glutamine synthetase through promoter demethylation. Cell Metab 22(6): 1068–1077.
Vincent EE, Sergushichev A, Griss T, Gingras M-C, Samborska B, Ntimbane T, Coelho PP, Blagih J, Raissi T, Choinière L, Bridon G, Loginicheva E, Flynn BR, Thomas EC, Tavaré JM, Avizonis D, Pause A, Elder DJ, Artyomov MN, Jones RG. 2015. Mitochondrial phosphoenolpyruvate carboxykinase (PCK2) regulates metabolic adaptation and enables glucose-independent tumor growth. Mol Cell 60(2):195–207.
Dupuy F, Tabariès S, Andrzejewski S, Dong Z, Blagih J, Annis MG, Omeroglu A, Gao D, Leung S, Amir E, Clemons M, Aguilar-Mahecha A, Basik M, Vincent EE, St.-Pierre J, Jones RG, Siegel PM. 2015. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab 22(4):577–589.
Fu A, Robitaille K, Faubert B, Reeks C, Dai X-X, Hardy A, Sankar A, Rocheleau JV, Wheeler MB, MacDonald PE, Jones RG, Screaton RA. 2015. LKB1 couples glucose metabolism to insulin secretion. Diabetologia 58(7):1513–1522.
Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, DeBerardinis RJ, Feron O, Frezza C, Ghesquiere B, Gottlieb E, Hiller K, Jones RG, Kamphorst JJ, Kibbey RG, Kimmelman AC, Locasale JW, Lunt SY, Maddocks O, Malloy C, Metallo CM, Meuillet EJ, Munger J, Nöh K, Rabinowitz JD, Ralser M, Sauer U, Stephanopoulos G, St.-Pierre J, Tennant DA, Wittmann C, Vander Heiden MG, Vazquez A, Vousden K, Young JD, Zamboni N, Fendt S-M. 2015. A roadmap for interpreting 13C metabolite labeling patterns from cells. Current Opin Biotechnol 34:189–201.
Belle JI, Langlais D, Petrov JC, Pardo M, Jones RG, Gros P, Nijnik A. 2015. p53 mediates loss of hematopoietic stem cell function and lymphopenia in Mysm1-deficiency. Blood 125(15):2344–2348.
Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vazquez G, Yurchenko E, Raissi TC, van der Windt GJW, Viollet B, Pearce EL, Pelletier J, Piccirillo CA, Krawczyk CM, Divangahi M, Jones RG. 2015. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42(1):41–54.
Vincent EE, Coelho PP, Blagih J, Griss T, Viollet B, Jones RG. 2015. Differential effects of AMPK agonists on cell growth and metabolism. Oncogene 34(28):3627–3639.
Poffenberger MC, Jones RG. 2014. Amino acids fuel T cell-mediated inflammation. Immunity 40(5):635– 637.
Yan M, Gingras MC, Dunlop EA, Nouët Y, Dupuy F, Jalali Z, Possik E, Coull BJ, Kharitidi D, Dydensborg AB, Faubert B, Kamps M, Sabourin S, Preston RS, Davies RS, Roughead T, Chotard L, van Steensel MA, Jones RG, Tee AR, Pause A. 2014. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J Clin Invest 124(6):2640–2650.
Possik E, Jalali Z, Nouët Y, Yan M, Gingras M-C, Schmeisser K, Panaite L, Kharitidi D, Dupuy F, Chotard L, Jones RG, Hall DH, Pause A. 2014. Folliculin regulates AMPK-dependent autophagy and metabolic stress survival. PLoS Genet 10(4):e1004273.
Wynosky-Dolfi MA, Snyder AG, Philip NH, Doonan PJ, Poffenberger MC, Avizonis D, Zwack EE, Riblett AM, Hu B, Strowig T, Flavell RA, Jones RG, Freedman BD, Brodsky IE. 2014. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J Exp Med 211(4):653–668.
Everts B, Amiel E, Huang SC, Smith AM, Chang C-H, Lam WY, Redmann V, Frietas TC, Blagih J, van der Windt GJ, Artyomov MN, Jones RG, Pearce EL, Pearce EJ. 2014. TLR-driven early glycolytic reprogramming via TBK1-IKK the anabolic demands of dendritic cell activation. Nat Immunol 15(4):323–332.
Faubert B, Vincent EE, Poffenberger MC, Jones RG. 2014. The AMP-activated protein kinase (AMPK) and cancer: Many faces of a metabolic regulator. Cancer Lett. 356(2):165–170.
Faubert B, Vincent EE, Griss T, Svensson R, Mamer OA, Avizonis D, Shaw RJ, Jones RG. 2014. Loss of LKB1 promotes metabolic reprogramming of cancer cells via HIF1. Proc Nat Acad Sci U S A 111(7):2554–2559.
McGuirk S, Gravel S-P, Deblois G, Papadopoli DJ, Faubert B, Wegner A, Hiller K, Avizonis D, Akavia UD, Jones RG, Giguère V, St.-Pierre J. 2013. PGC-1alpha supports glutamine metabolism in breast cancer. Cancer Metab 1(1):22.
Pearce EL, Poffenberger M, Chang C-H, Jones RG. 2013. Fueling immunity: New insights on metabolism and lymphocyte function. Science 342(6155):1242454.
Dupuy F, Dong Z, Avizonis D, Ling C, Griss T, Siwak DR, Mills GB, Muller WJ, Siegel PM, Jones RG. 2013. LKB1 loss promotes ErbB2-driven breast cancer development but impairs the growth of lung metastases. Cancer Metab 1(1):18.
van der Windt GJW, O’Sullivan D, Everts B, Huang SC-C, Buck MD, Curtis JD, Chang C-H, Smith AM, Ai T, Faubert B, Jones RG, Pearce EJ, Pearce EL. 2013. Bioenergetic differences underlie the ‘rapid recall’ capability of CD8 memory T cells. Proc Nat Acad Sci U S A 110(35):14336–14341.
Chang C-H, Curtis JD, Maggi Jr LB, Faubert B, Villarino AV, O’Sullivan D, Huang SC-C, van der Windt GJW, Blagih J, Qui J, Weber JD, Pearce EJ, Jones RG, Pearce EL. 2013. Post-transcriptional control of T Cell effector function by aerobic glycolysis. Cell 153(6):1239–1251.
Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo A, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, Faubert B, Bridon G, Tognon CE, Mathers J, Thomas R, Li A, Barokas A, Bowden M, Smith S, Wu X, Korshunov A, Hielscher T, Northcott PA, Galpin JD, Ahern CA, Wang Y, McCabe MG, Collins P, Jones RG, Pollak MN, Delattre O, Gleave ME, Jan E, Pfister SM, Proud CG, Derry WB, Taylor MD, Sorensen PHB. 2013. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153(5):1064–1079.
O’Shea JJ, Jones RG. 2013. Autoimmunity: Rubbing salt in the wound. Nature 496(7446):437–439.
Sanchez-Macedo N, Feng J, Faubert B, Chang N, Elia A, Rushing EJ, Tsuchihara K, Bungard D, Berger SL, Jones RG, Mak TW, Zaugg K. 2013. Depletion of the novel p53-target gene carnitine palmitoyltransferase 1C delays tumor growth in the neurofibromatosis type I tumor model. Cell Death Differ 20(4):659–658.
Delisle J-S, Giroux M, Boucher G, Landry J-R, Hardy M-P, Lemieux S, Jones RG, Wilhelm BT, Perreault C. 2013. The TGF-/Smad3 pathway inhibits CD28-dependent cell growth and proliferation of CD4 T cells. Genes Immun 14(2):115–126.
Faubert B, Boily B, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, Mamer OA, Avizonis D, DeBerardinis RJ, Siegel PM, Jones RG. 2013. AMPK is a negative regulator of the Warburg Effect and suppresses tumor growth in vivo. Cell Metab 17(1):113–124.
Christie DA, Mitsopoulos P, Blagih J, Dunn SD, St. Pierre J, Jones RG, Hatch GM, Madrenas J. 2012. SLP-2 deficiency in T cells is associated with impaired mitochondrial respiration and defective CD4+ T cell responses. J Immunol 189(9):4349–4360.
Blagih J, Krawczyk CM, Jones RG. 2012. LKB1 and AMPK: Central regulators of lymphocyte metabolism and function. Immunol Rev 249(1):59–71.
Blagih J, Jones RG. 2012. Polarizing macrophages through reprogramming of glucose metabolism. Cell Metab 15(6):793–795.
MacIver N, Blagih J, Saucillo DC, Tonelli L, Griss T, Rathmell JC, Jones RG. 2011. The liver kinase B-1 (LKB1) is a central regulator of T cell development, activation, and metabolism. J Immunol 187(8):4187–4198.
Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B, Kalliomaki T, Elia A, Luo X, Nadeem V, Bungard D, Yalavarthi S, Growney JD, Wakeham A, Moolani Y, Silvester J, Ten AY, Bakker W, Tsuchihara K, Berger SL, Hill RP, Jones RG, Tsao M, Robinson MO, Thompson CB, Pan JG, Mak TW. 2011. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev 25(10):1041–1051.
Bungard D, Fuerth B, Yang PY, Faubert B, Mass NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. 2010. Signaling kinase AMPK regulates stress-promoted transcription via Histone H2B phosphorylation. Science 329(5996):1201–1205.
Moser TS, Jones RG, Thompson CB, Coyne CB, Cherry S. 2010. A kinome RNAi screen identified AMPK as promoting poxvirus entry through the control of actin dynamics. PLoS Pathog. 6(6):e1000954.
Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, Cross JR, Jung E, Thompson CB, Jones RG, Pearce EJ. 2010. Toll-like receptor induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115(23):4742–4749.
Jones RG. 2009. The roles, mechanisms, and controversies of autophagy in mammalian biology. F1000 Biology Reports 1:68.
Jones RG, Thompson CB. 2009. Tumor suppressors and cancer metabolism: a recipe for cancer growth. Genes Dev 23(5):537–548.
Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. 2009. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460(7251):103–107.
Jones RG, Thompson CB. 2007. Revving the engine: Signal transduction fuels T cell activation. Immunity 27(2):173–178.
Jones RG, Bui T, White C, Madesh M, Krawczyk CM, Lindsten T, Hawkins BJ, Kubek S, Frauwirth KA, Wang YL, Conway SJ, Roderick HL, Bootman MD, Shen H, Foskett JK, Thompson CB. 2007. The proapoptotic factors Bax and Bak regulate T cell proliferation through control of endoplasmic reticulum Ca2+ homeostasis. Immunity 27(2):268–280.
Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. 2007. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res 67(14):6745–6752.
Saibil SD, Jones RG, Deenick EK, Liadis N, Elford AR, Vainberg MG, Baerg H, Woodgett JR, Gerondakis S, Ohashi PS. 2007. CD4+ and CD8+ T cell survival is regulated differentially by PKC, c-Rel and PKB. J Immunol 178(5):2932–2939.
Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. 2006. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell 21:521–531.
Jones RG, Saibil SD, Pun JM, Elford AR, Bonnard M, Pellegrini M, Arya S, Parsons MR, Krawczyk CM, Gerondakis S, Yeh W-C, Woodgett JR, Boothby MR, Ohashi PS. 2005. NF-kB couples protein kinase B/Akt signaling to distinct survival pathways and the regulation of lymphocyte homeostasis in vivo. J Immunol 175:3790–3799.
Buzzai M, Bauer DE, Jones RG, DeBerardinis RJ, Hatzivassiliou G, Elstrom RL, Thompson CB. 2005. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid b-oxidation. Oncogene 24(26):4165–4173.
Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18(3):283–293.
Krawczyk CM, Jones RG, Atfield A, Bachmaier K, Arya S, Odermatt B, Ohashi PS, Penninger JM. 2005. Differential control of CD28-regulated in vivo immunity by the E3 ligase Cbl-b. J Immunol 174(3):1472–1478.
Jeon MS, Atfield A, Venuprasad K, Krawczyk CM, Sarao R, Elly C, Yang C, Arya S, Bachmaier K, Su L, Bouchard D, Jones RG, Gronski M, Ohashi P, Wada T, Bloom D, Fathman CG, Liu YC, Penninger JM. 2004. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity 21(2):167–177.
Berg-Brown NN, Gronski MA, Jones RG, Elford AR, Deenick EK, Odermatt B, Littman DR, Ohashi PS. 2004. PKC signals activation versus tolerance in vivo. J Exp Med 199(6):743–752.
Jones RG. 2003. CD28 signaling and T cell death. Mod Asp Immunobiol 3(1):4–5.
Jones RG, Elford AE, Parsons MJ, Wu L, Krawczyk CM, Yeh W-C, Hakem R, Rottapel R, Woodgett JR, Ohashi PS. 2002. CD28-dependent activation of protein kinase B/Akt blocks Fas-mediated apoptosis by preventing death-inducing signaling complex assembly. J Exp Med 196(3):335–348.
Parsons MJ, Jones RG, Tsao M-S, Odermatt B, Woodgett JR, Ohashi PS. 2001. Expression of active protein kinase B in T cells perturbs both T and B cell homeostasis and promotes inflammation. J Immunol 167(1):42–48.
Garza KM, Nguyen LT, Jones RG, Ohashi PS. 2001. Factors contributing to autoimmune disease. Adv Exp Med Biol 490:7–19.
Krawczyk CM, Bachmaier K, Sasaki T, Jones RG, Snapper SB, Bouchard D, Kozieradzki I, Ohashi PS, Alt FW, Penninger JM. 2000. Cbl-b is a negative regulator of receptor clustering and raft aggregation in T cells. Immunity 13(4):463–473.
Ohteki T, Parsons M, Zakarian A, Jones RG, Nguyen LT, Woodgett JR, Ohashi PS. 2000. Negative regulation of T cell proliferation and interleukin 2 production by the serine threonine kinase GSK-3. J Exp Med 192(1):99–104.
Jones RG, Parsons M, Bonnard M, Chan VS, Yeh WC, Woodgett JR, Ohashi PS. 2000. Protein kinase B regulates T lymphocyte survival, nuclear factor kappaB activation, and Bcl-X(L) levels in vivo. J Exp Med 191(10):1721–1734.
Mariathasan S, Jones RG, Ohashi PS. 1999. Signals involved in thymocyte positive and negative selection. Semin Immunol 11(4):263–272.
Sebzda E, Mariathasan S, Ohteki T, Jones RG, Bachmann MF, Ohashi PS. 1999. Selection of the T cell repertoire. Annu Rev Immunol 17:829–874.
Sasaki T, Irie-Sasaki J, Jones RG, Oliveira dos Santos AJ, Stanford WL, Bolon B, Wakeham A, Itie A, Bouchard D, Kozieradzki I, Joza N, Mak TW, Ohashi PS, Suzuki A, Penninger JM. 1999. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science 287(5455): 1040–1046.
Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. 1998. X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science 280(5367):1277–1281.

Michael Dahabieh, Ph.D.
Postdoctoral Fellow, Russell Jones Laboratory
Characterizing heterogeneity of exhausted T cells and understanding the roles of peroxisomes in the tumor microenvironment
Lisa DeCamp
Laboratory Manager, Department of Metabolism and Nutritional Programming

Nicole Foy
Assistant Research Technician, Department of Metabolism and Nutritional Programming

Susan Kitchen-Goosen, B.S., MBA
Research Associate, Department of Metabolism and Nutritional Programming

Dahlya Kamarudin, M.S.
Ph.D. Student, VAI Graduate School
Research focus to be determined.

Brandon Oswald, Ph.D.
Postdoctoral Fellow, Russell Jones Laboratory
Thesis: Caloric restriction in mice increases the functionality of T cells, offsetting tumor burden

Ilaria Panzeri, Ph.D.
Research Scientist, Department of Metabolism and Nutritional Programming
Epigenetic origins of phenotypic variation in complex metabolic diseases


Jeanie Wedberg
Senior Administrative Assistant II, Department of Metabolism and Nutritional Programming

Kelsey Williams, Ph.D.
Research Program Manager, MeNu


